Novel computational tools and genomic resources have brought a molecular perspective on the Hydra stem cell system. A major breakthrough in research on cnidarian development came with the generation of stably transgenic Hydra by the Bosch lab using embryo injection (Wittlieb et al., 2006).
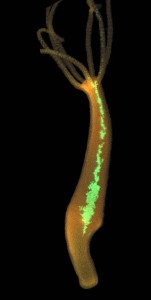
This opened up a new range of experimental possibilities for studying regeneration and cellular behavior in vivo in this model organism (Siebert et al., 2008; Wittlieb et al., 2006; Khalturin et al., 2007; Gee et al., 2010; Nakamura et al., 2011; Böhm et al., 2012; Franzenburg et al., 2012) indicating the value of this approach. The successful creation of transgenic Hydra expressing GFP in all three stem cell lineages (Hemmrich et al., 2012) has greatly facilitated progress in getting insights of general relevance into stem cell biology including cellular senescence, lineage programming, the role of extrinsic signals in fate determination and tissue homeostasis, and the evolutionary origin of these cells. Loss-of-function approaches based on hairpin constructs have been successfully used to knockdown both immune (Franzenburg et al., PNAS 2012, PNAS 2013) as well as stem cell transcripts (Boehm et al., PNAS 2012).
We also provide a comparative genomics platform (http://www.compagen.org/) which provides access to a large set of genome and transcriptome sequences from basal metazoans including sponges and Cnidaria species. Since the genome organization and genome content of Cnidaria is remarkably similar to that of bilaterians, these animals offer unique insights into the content of the “genetic tool kit” present in the Cnidarian–bilaterian ancestor.
